|
|
|
-
To export sequences to Fasta format, click on File to go to the backstage section and select Export.
-
Choose on top, which records to be included in this export:
-
Export all records This will export all records in the current table view.
-
Export selected records only This will take only the selected records in the current table view (with the Shift or Ctrl keys pressed for multiple selection).
-
-
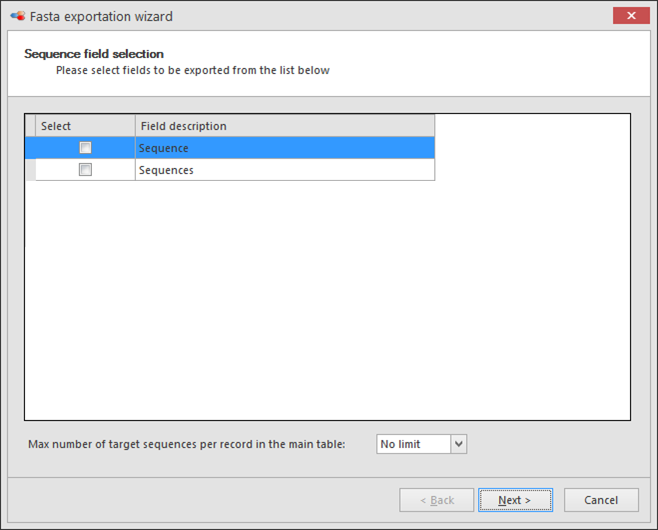
Check the sequence field(s) to be exported. Note that all N and NLink fields of the current table are listed.
When multiple sequences are linked to a record by an NLink field, then set Max number of target sequences per record in the main table to be exported (choose from 1, 2, 5, 10, 20, 50, 100 or no limit).
As an example, when the option '1' target record is chosen, only the first linked record in the list will be exported. When the option '5' target records is chosen, the first 5 linked records will be exported, per record and per NLink field. Therefore, if one selects only one sequence record to be exported, the "good" one must be the first in the list of the linked sequence records. To change the order of the linked records see Links management, buttons 6-9.
Note, multiple fields can be selected, right click, Select highlighted fields.
Click Next to go to the next step.
-
Select the field(s) to be included in the header/title of the sequences in the exported Fasta file (optional).
Note, multiple fields can be selected, right click, Select highlighted fields.
Click Next to go to the next step.
-
Select the directory to save the Fasta file. BioloMICS will save it automatically to the correct directory where local blast alignments will be performed. To save it at another location, click Browse.
Check Format the file for BLAST alignment to automatically create three files needed for performing a BLAST using this export as a local reference database.
Check Export sequences with short titles to export only the Name of the record in the grid, the name of the linked sequence and the selected additional fields of step 5 above.
Click Next to go to the next step and to Finish the wizard.
|
|
|
|
 Note, multiple fields can be selected, right click, Select highlighted fields.Click Next to go to the next step.
Note, multiple fields can be selected, right click, Select highlighted fields.Click Next to go to the next step.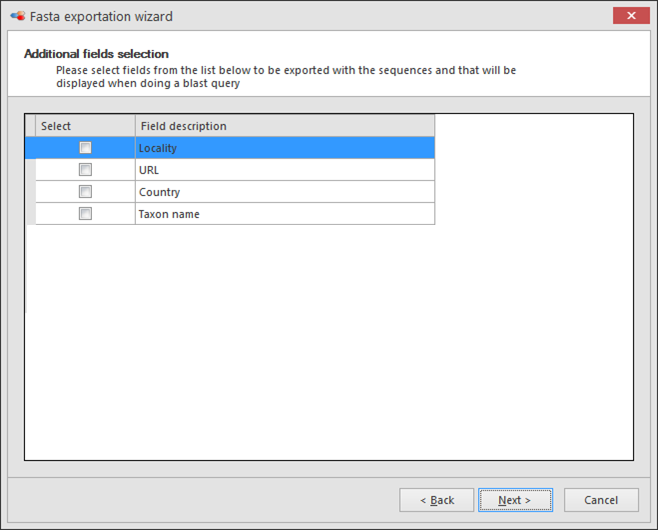 Note, multiple fields can be selected, right click, Select highlighted fields.Click Next to go to the next step.
Note, multiple fields can be selected, right click, Select highlighted fields.Click Next to go to the next step.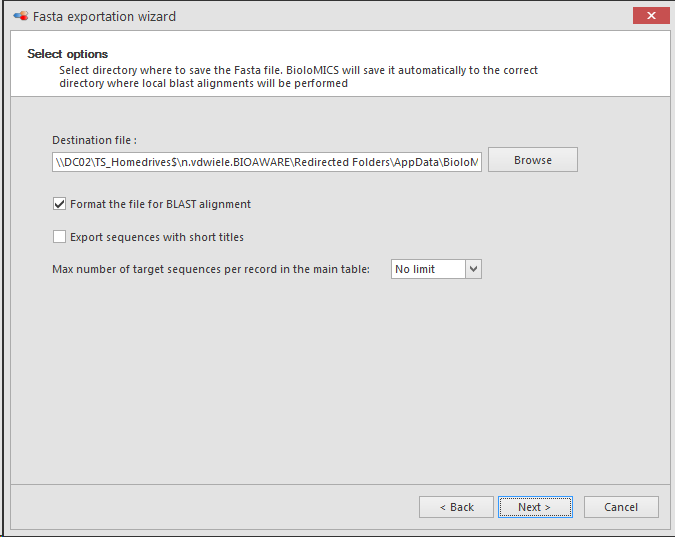 Click Next to go to the next step and to Finish the wizard.
Click Next to go to the next step and to Finish the wizard.
