BLAST / Align sequences with reference db
-
Select the records in the BioSheet, to be used in the pairwise alignment (with the Shift or Ctrl keys pressed for multiple selection).
-
To align multiple sequences, in the Ribbon of the main window of BioloMICS, under Molecular Tools, in the Pairwise alignment group, click Align sequences with reference DB.

-
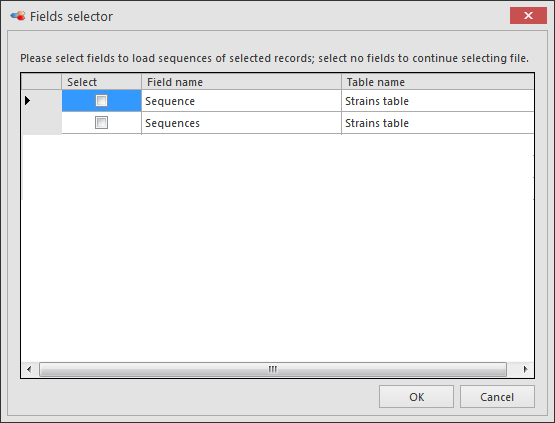
Select the sequence field(s) to be included in this pairwise alignment and click OK.
-
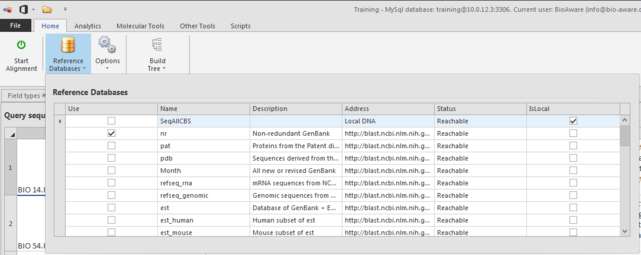
Click Reference databases and select all the wanted reference databases for this pairwise alignment.
-
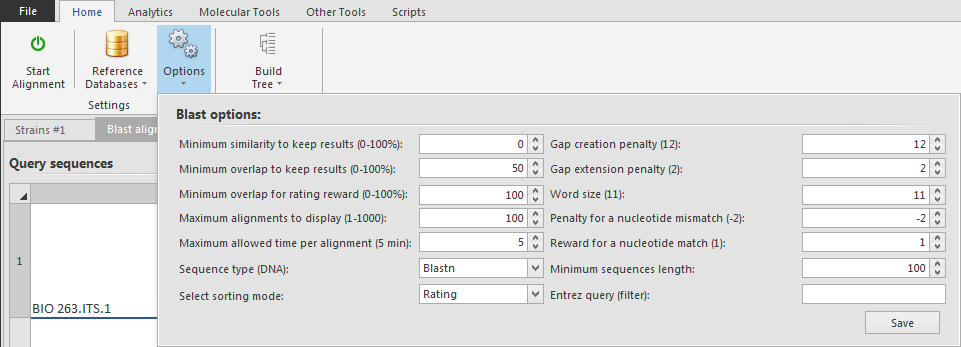
Click Options to set the Blast options for this pairwise alignment. Click Save to save the changes.
-
Entrez query (filter) A BLAST search van be limited to the rsult of an Entrez query against the database chose. This restricts the search to a subset of entries from that database fitting the requirement of the Entrez query. Terms normally accepted by Entrez nucleotide or protein searches are accepted here. Examples are given below.
-
protease NOT hiv1[organism]This will limit a BLAST search to all proteases, except those in HIV 1.
-
1000:2000[slen]This limits the search to entries with lengths between 1000 to 2000 bases for nucleotide entries, or 1000 to 2000 residues for protein entries.
-
Mus musculus[organism] AND biomol_mrna[properties]This limits the search to mouse mRNA entries in the database. For common organisms, one can also select from the pulldown menu.
-
10000:100000[mlwt]This is yet another example usage, which limits the search to protein sequences with calculated molecular weigt between 10 kD to 100 kD.
-
src specmin voucher[properties]This limits the search to entries that are annotated with a /specimen)voucher quilifier on the dource feature.
-
all[filter] NOT environmental sample[filter] NOT metagenomes[orgn]
-
all[filter] NOT uncultured[filter] NOT environmental sample[filter] sequence from type [filter]This excludes sequences from metagenome studies and uncultured sequences from anonymous environmental sample studies.For help in constructing Entrez queries please see the "Writing Advanced Search Statements <http://www.ncbi.nlm.nih.gov/books/NBK3837/>" section of the Entrez Help document. Knowing the content of a database and applying Entrez terms accordingly are important. For example, biomol_mrna[prop] should not be applied to htgs or chromosome database since they do not contain mRNA entries!
-
-
Click Start alignment to start the pairwise alignment and to see the results.

-
Select one of the sequences in the top grid to see the best hits in the lower-left grid.The lower-right textbox shows the alignment of the selected selected sequence in the top grid and the selected alignment result in the lower-left grid.
-
When Genbank is used as a reference, select the Genbank hit in the alignment results and click on the cell in the URL column to open the Genbank website of the selected hit.
This movie shows how to blast versus local and external databases in BioloMICS.
1. Sequence fields (N & NLink) (0:10)
2. Create local reference file (0:21)
3. Export sequences to Fasta (0:28)
4. Molecular tools - Align Sequences with Reference db (1:26)
5. Select reference databases (1:49)
6. Set Blast options (2:11)
7. Start alignment (2:19)
8. Results (2:25)

