Use pairwise alignment (user)
Visitors of the website can perform an alignment based on the reference databases the admin made available.
NB. The alignment template was created by the administrator of the website and can be used by the user. For instructions on how to create the alignment template, see Create / activate pairwise alignment (admin).
-
Go to the website and navigate to the alignment page.
-
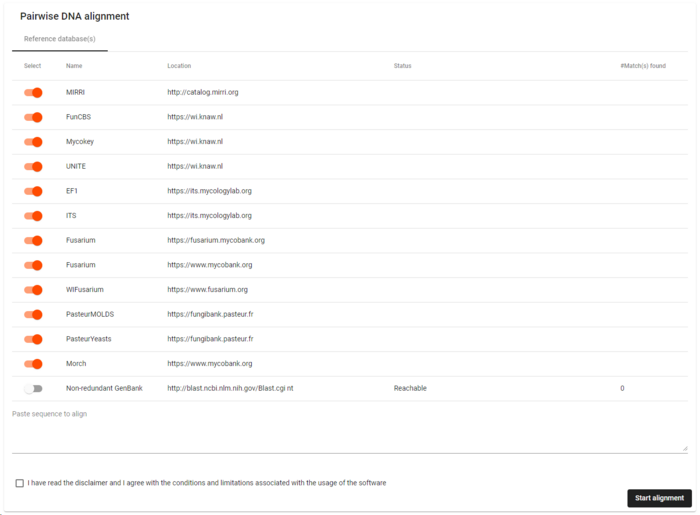
Enable the reference database(s) to be used for this analysis.
-
Enter the sequences that will be compared to the sequences in the reference database(s).
-
Read the disclaimer and check the box to agree.
-
Click Start alignment.
-
The results are shown below, the top result is the best match.
The following instruction movie explains in detail how to perform a pairwise alignment on the website:
1. Reference databases to be selected (0:08)
2. Paste sequence (0:18)
3. Read and accept the disclaimer (0:22)
4. Read and accept disclaimer (0:40)
5. Start alignment (0:30)
6. Show results (0:48)
7. Show alignment (0:52)
8. Draw tree (0:59)
The pairwise alignment tool can be activated by the administrator of the website.
Instructions on how to establish the wanted criteria, see chapter Alignment or Create / activate pairwise alignment.
Once the right reference databases are added to the template alignment the user can:
-
Navigate to the specific web page: WebsiteName/page/PageName.
-
Select the reference databases to be used in this alignment and deselect the ones that should not be used.
-
Paste the sequence to be compared.
-
Read, and check the box "I have read the disclaimer and I agree with the conditions and limitations associated with the usage of the software".
-
Click on the button Start alignment.
-
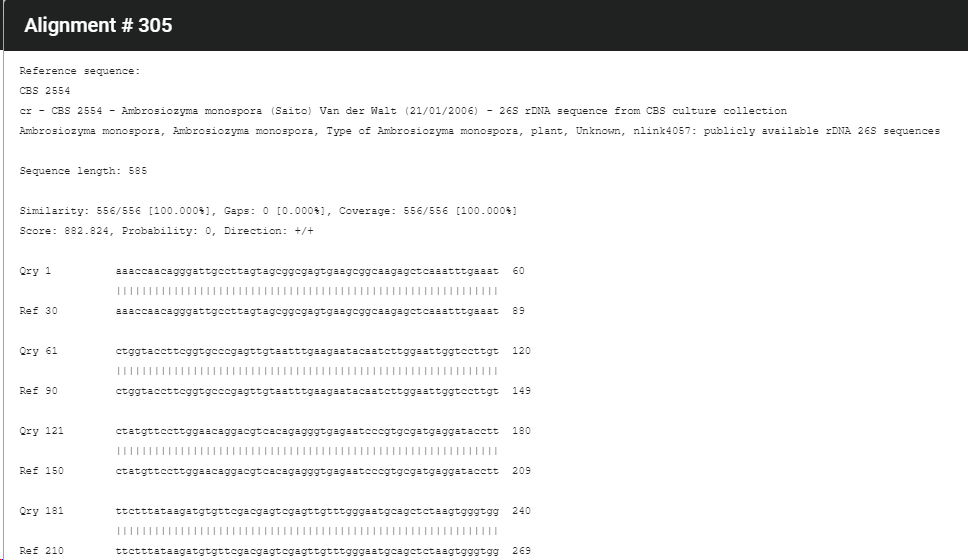
The results will be displayed. The best results is shown at the top of the list.
-
Click on the button Show alignments to see more information about the compared sequences.
-
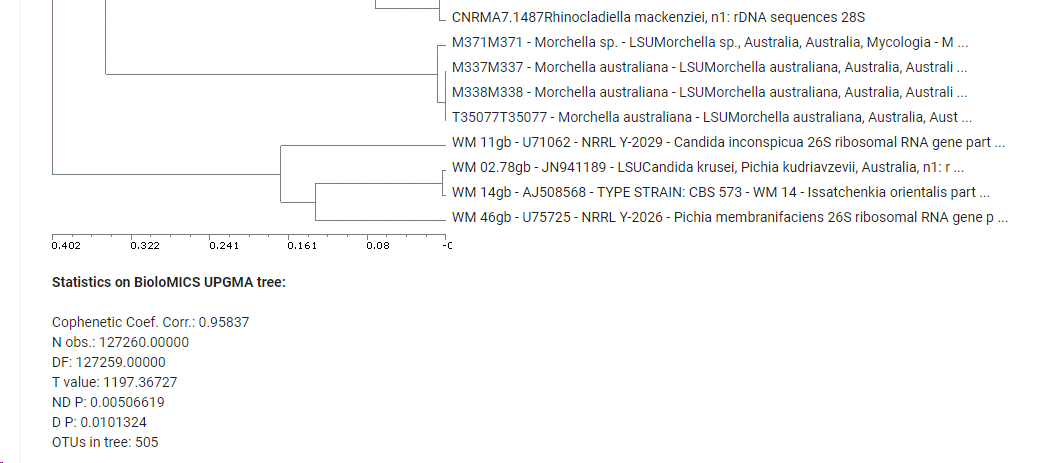
Click Draw tree to build a tree based on the results.
-
Choose the wanted algorithm: UPGMA, WPGMA, Single linkage, Complete linkage, Neighbor joining, Ward, UPGMC, WPGMC.