Multiple Alignment
The multiple alignment is a sequence alignment of three or more sequences. In many cases, the input set of sequences is assumed to have an evolutionary relationship.
BioloMICS uses a multiple sequence alignment by log-expectation method. The distance measure is updated between iteration stages.
Six different strategies for alignments are available. They will define the accuracy and the time to process the sequences.
To choose multiple alignment strategy, click on the More button and select Settings. In the Options dialog go to Alignments and set the strategy under Multi alignments.
-
Select the records in the BioSheet, to be used in the multiple alignment (with the Shift or Ctrl keys pressed for multiple selection).
-
In the Ribbon of the main window of BioloMICS, under Molecular Tools, in the Multiple alignment group, click Multiple alignment of selected records.
 Note that when no records were selected from the BioSheet, then a popup appears and externally saved sequences can be loaded.
Note that when no records were selected from the BioSheet, then a popup appears and externally saved sequences can be loaded. -
Select the sequence field(s) to be included in this multiple alignment and click OK.
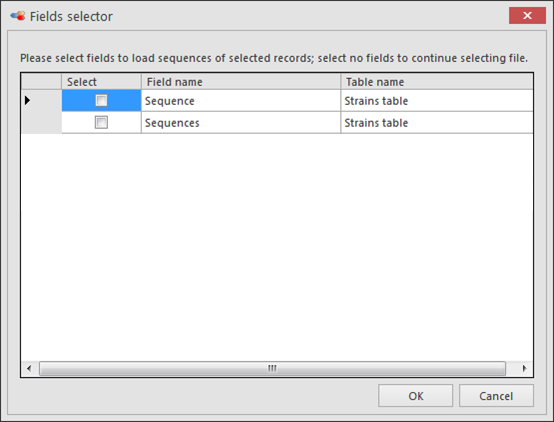 Note that when no field is selected, then a popup appears and externally saved sequences can be loaded.
Note that when no field is selected, then a popup appears and externally saved sequences can be loaded. -
The sequence viewer will be opened:
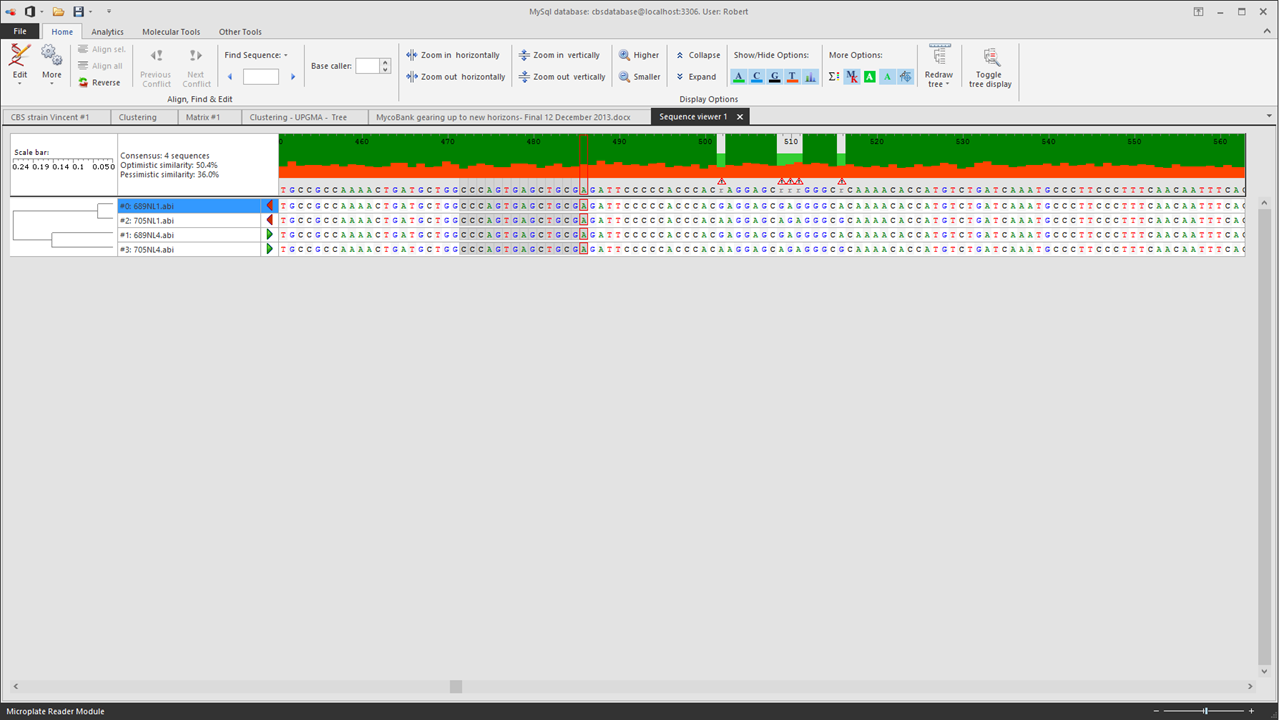
-
For more details on editing see Sequence edition and Ribbon multiple alignment and trace file edition.
-
To save your sequences, click on the save button
 at the top-left of the window and choose from one of the following options:
at the top-left of the window and choose from one of the following options: -
Save (as) ... to import the sequences into the database. More...
-

