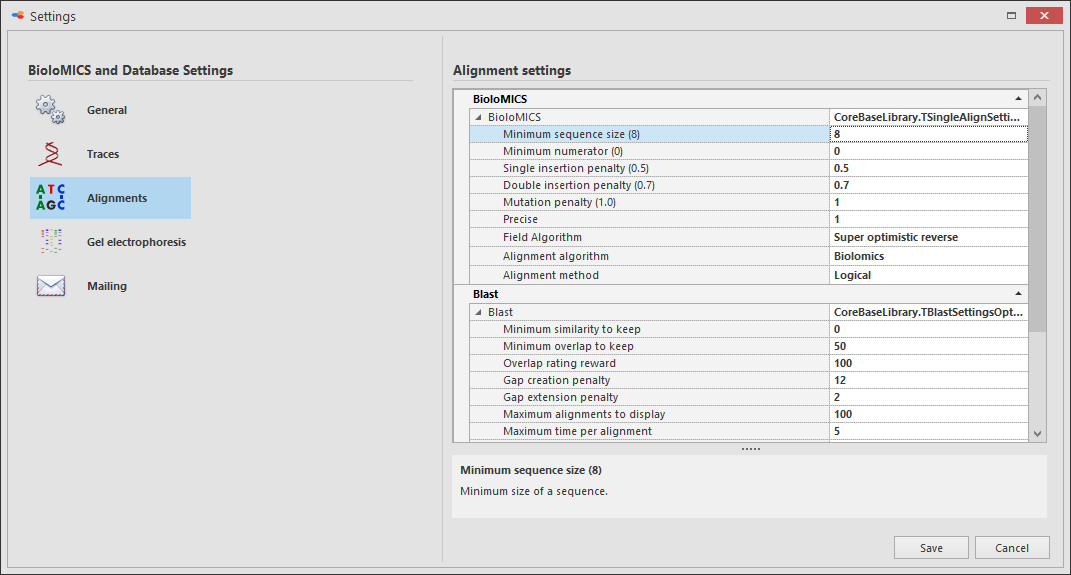
Alignments
BioloMICS:
-
Minimum sequence size (8) Minimum size of a sequence.
-
Minimum numerator (0) Minimum numerator (0).
-
Single insertion penalty (0.5) Penalty for a single insertion.
-
Double insertion penalty (0.7) Penalty for a double insertion.
-
Mutation penalty (1.0) Penalty for a mutation.
-
Precise Precise.
-
Field algorithm Algorithms listed for different field types.
-
Alignment algorithm Biolomics, Blast, Fasta or MultiAlign.
-
Alignment method Logical, BestCase or ExactMatch.
Blast:
-
Minimum similarity to keep results Minimum similarity to keep results.
-
Minimum overlap to keep results Minimum overlap to keep results.
-
Overlap rating reward Overlap rating reward.
-
Gap creation penalty Penalty when creating a gap.
-
Gap extension penalty Penalty when extending a gap.
-
Maximum alignments to display Maximum number of alignments to display.
-
Maximum allowed time per alignment Maximum allowed time per alignment.
-
Minimum sequences length Minimum length of a sequence.
-
Penalty mismatch Penalty for a mismatch.
-
Reward match Reward for a match.
-
Word size Length of the initial words. Default is 11, but can be decreased (to a minimum of 7) to increase the sensitivity. When increasing the word size, the search speed will be increased.
-
Sequence type Blastn, Blastp, Blastx, TBlastn, TBlastx, Fastan or Fastap.
-
Select sorting mode Rating, Similarity or Probability.
Multiple alignments:
-
Strategy
-
Automatic detection of the most appropriate algorithm
-
Very fast progressive method with a rough guide tree
-
Fast progressive method
-
Accurate iterative method suitable for less than 200 sequences of similar length
-
Most accurate iterative refinement method suitable for less than 200 sequences
-
Accurate method suitable for less than 200 sequences containing large unalignable regions
Click Save to save the changes made and to close the Options dialog.