Global options Alignment
Click on a reference in the template area to see the properties panel on the right.
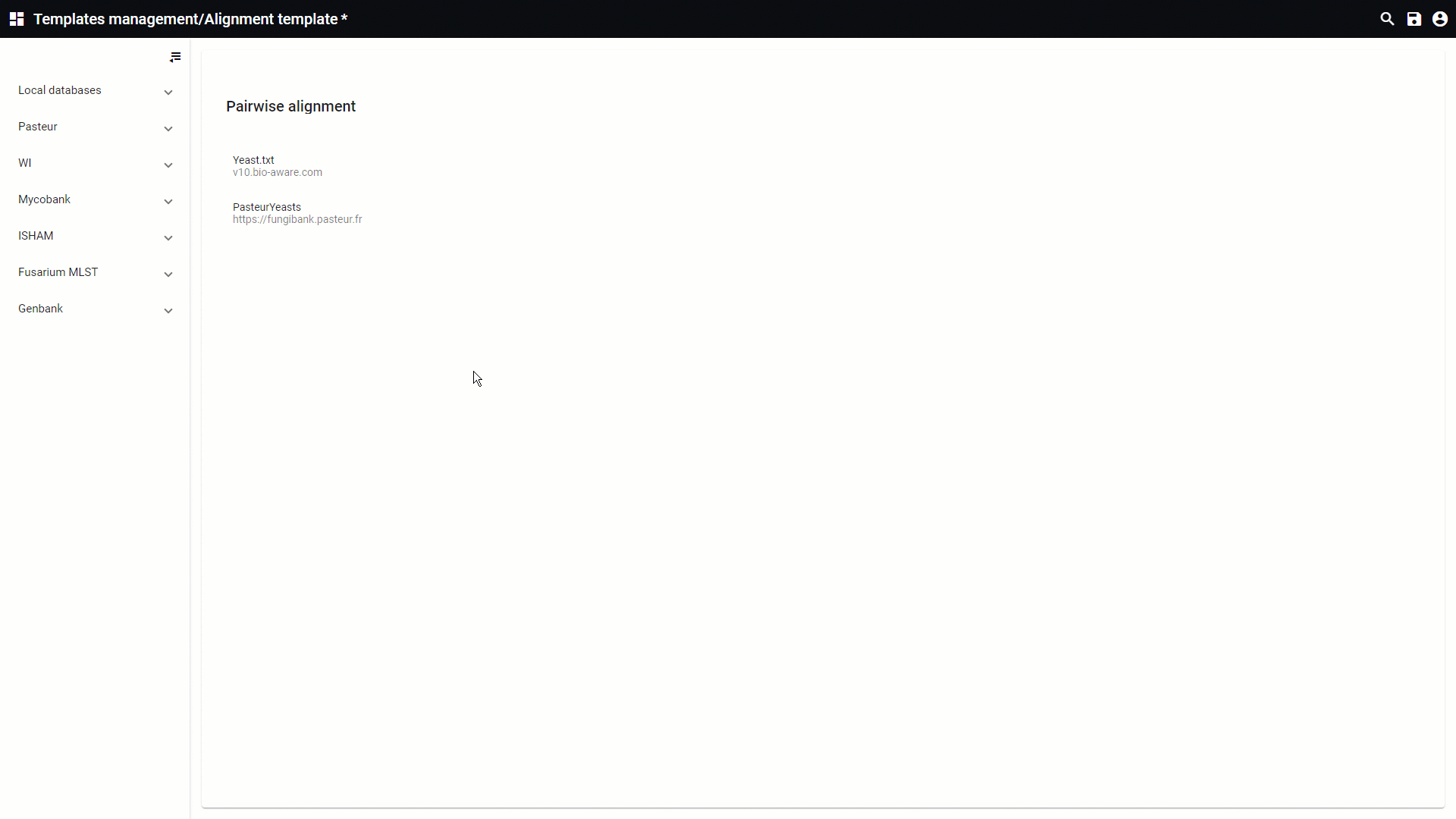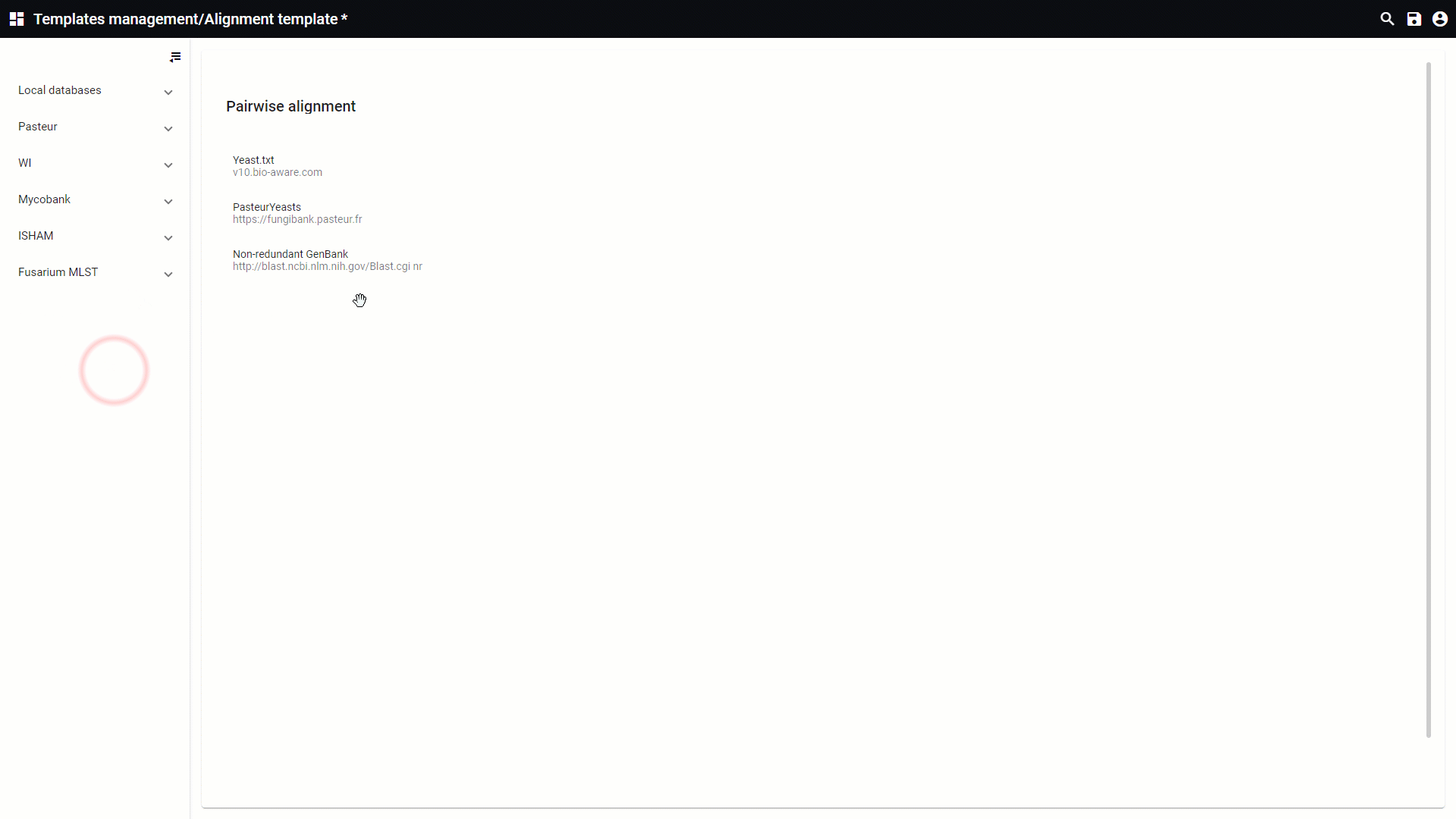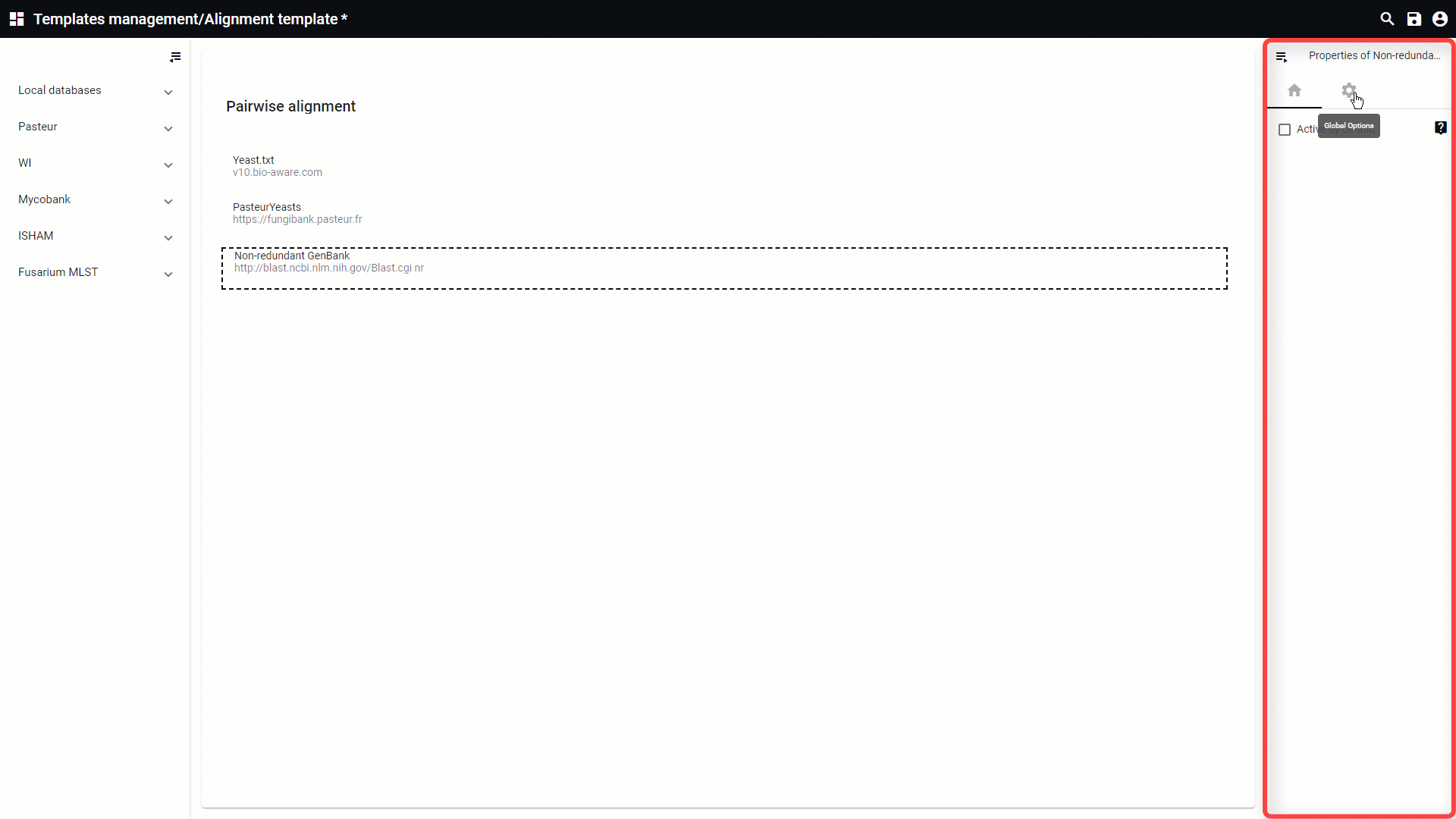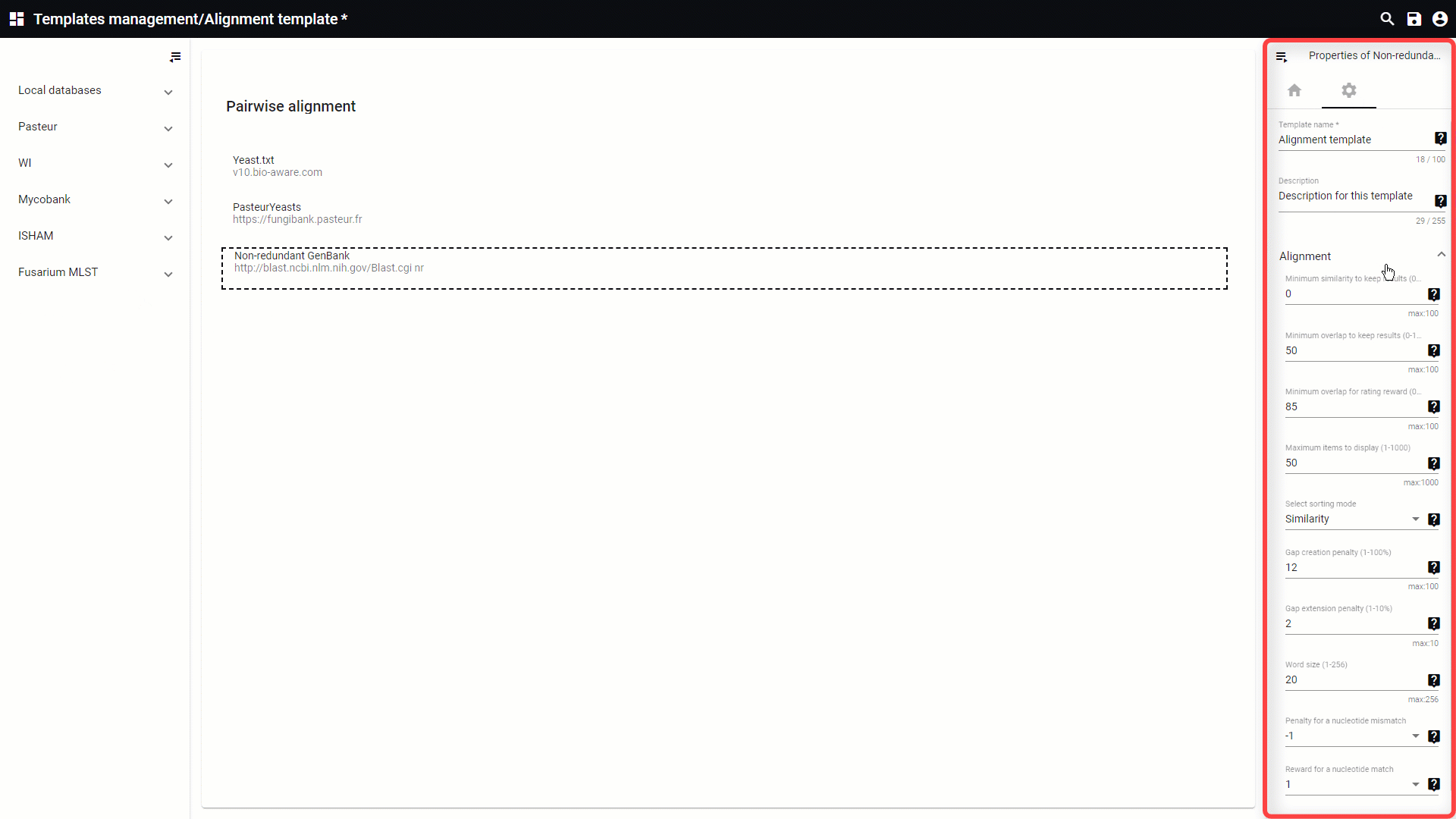
-
Minimum similarity to keep results (0-100%): Filters data on similarity rate (%).
-
Minimum overlap to keep results (0-100%): Minimum overlap needed before the results are displayed.
-
Minimum overlap for rating reward (0-100%): This value specifies the required overlap (%) to get a rating reward.
-
Maximum items to display (1-1000): This will limit the output to the specifies amount of alignments to display.
-
Select sorting mode:
-
Rating:***** When probability, similarity fragments and overlap matches (similarity = 100% 2*).**** When five of the upper conditions match (similarity >=95% only 1*).*** When three of the upper conditions match.** When two of the upper conditions match.* When only one of the upper conditions matches.
-
Similarity: This sorting takes only the similarity into account.
-
Probability: This sorting takes only the probability into account.
-
Gap creation penalty (1-100%): Penalty to open or extend a new gap in an alignment.
-
Gap extension penalty (1-10%): Penalty for gap extension (default is 2).
-
Word size (1-256): Used to nucleate regions of similarity. By default, it is 11 bp for nucleotide sequences, but can be decreased to a minimum of 7. When increasing the word size, the search speed will be increased.
-
Penalty for a nucleotide mismatch: When blasting the penalties are given when there is a nucleotide mismatch.
-
Reward for a nucleotide match: When blasting the scores are the reward for a nucleotide match.

