|
|
|
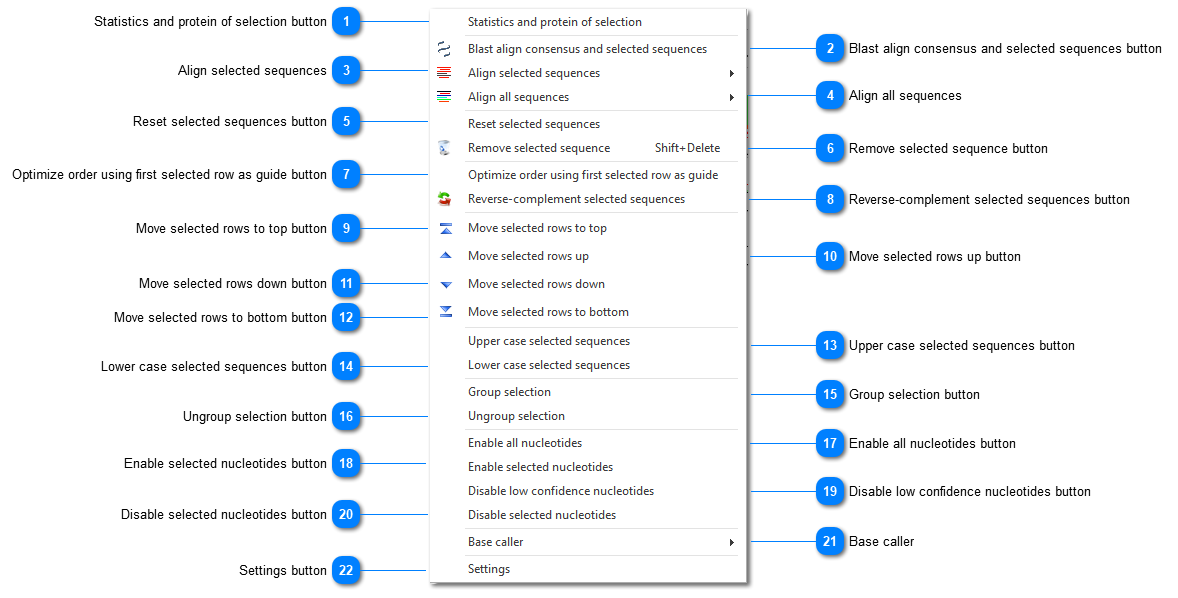
 Statistics and protein of selection buttonShow sequence statistics.
| |
 Blast align consensus and selected sequences buttonPerform blast alignment with consensus and the selected sequence(s).
| |
 Align selected sequencesSelect Optimized order to align the selected sequences in forward and reverse-complement directions, displaying the best result only. The second column (between the name of the file and the actual sequence) displays a green arrow pointing to the right when the sequence is displayed in forward direction and a red arrow pointing to the left when the reverse complement is displayed.
Select Original order to align the selected sequences using the current displayed direction. If the sequence is already displayed in reverse-complement, the sequence is NOT reset to forward direction before alignment.
| |
 Align all sequencesThe same as explained above in point 3 (Align selected sequences), but now using all sequences displayed in the sequence viewer.
| |
 Reset selected sequences buttonReset the selected sequences in the forward direction. It removes all previous offset computed during alignment.
| |
 Remove selected sequence buttonRemove the selected sequence from the experiment. Warning: there is no undo!
| |
 Optimize order using first selected row as guide buttonIn a multiple alignment result, it is possible that many sequences are displayed in reverse direction. In some cases, one sequence must remain in the forward direction and should be used to force all other sequences to align to its own content. This options keeps the first sequence in the current direction and adapt all others.
| |
 Reverse-complement selected sequences buttonReplace the selected sequence by its reverse-complement content.
| |
 Move selected rows to top buttonMove selected rows to the top of the sequence viewer, which may be necessary before aligning with the first sequence as a guide.
| |
 Move selected rows up buttonMove selected rows one place up.
| |
 Move selected rows down buttonMove selected rows one place down.
| |
 Move selected rows to bottom buttonMove selected rows to the bottom of the sequence viewer.
| |
 Upper case selected sequences buttonSet the selected sequence completely to upper case. In general the sequences are written in uppercase and the changes made by hand are kept in lowercase.
| |
 Lower case selected sequences buttonSet the selected sequence completely to lower case. In general the sequences are written in uppercase and the changes made by hand are kept in lowercase.
| |
 Group selection buttonGroup sequences together. The advantage is that when a change is made to one of the sequences in the group, all the other sequences in the group are changed automatically too.
| |
 Ungroup selection buttonUngroup the grouped sequences.
| |
 Enable all nucleotides buttonEnable all nucleotides.
| |
 Enable selected nucleotides buttonEnable selected nucleotides.
| |
 Disable low confidence nucleotides buttonDisplay in gray all nucleotides with a confidence below the confidence level (set in the Options dialog). They are automatically excluded from alignment computations.
| |
 Disable selected nucleotides buttonDisable selected nucleotides.
| |
 Base callerEach nucleotide contains two different letters: the original nucleotide and the one computed from the trace wave. As different base-caller algorithms give different results, it may be necessary either to re-compute the most probable nucleotide, or to reset it to the original value.
Two algorithms are currently available in the Options dialog, the Average method and the Max value method. For more details on the algorithms click here.
| |
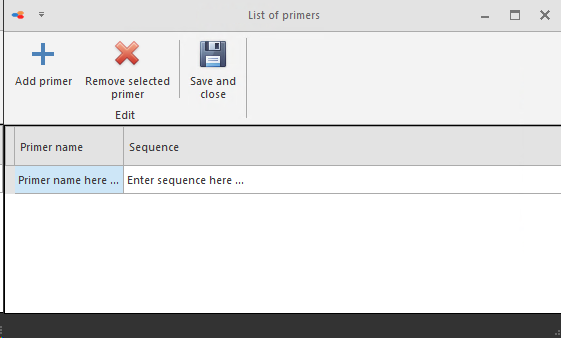
 Primers list
Primers can be added to the Primers list.
Click on the Add button and provide the Primer name and its sequence. Then click on the Save and close button.
To remove a primer from the list, click on the Remove selected primer button.
The primers in the list will be visible in a dropdown when clicking on the button Find primer.
| |
 Settings buttonOpens the Options dialog.
| |
|
|
|
|